Evaluation of the E3-ligase independent function of Transcription Intermediary Factor1γ (TIF1γ) in the regulation of cancer stem cells
SERB, Department of Science & Technology [DST]
Research Summary
Oral cancer is the second deadliest cancer in India. In 1 out of 2 patients, the disease comes back after treatment (recurrence). Self-renewing cancer cells or cancer stem cells (CSCs) are the reason for recurrence. My team was a part of a national initiative to study oral cancer in India. The leads we got from the study is being taken forward. Currently we are teaming up with PIs from RGCB and other BRIC institutes along with clinicians, to understand the molecular network that supports chemoresistance and recurrence in oral cancer. With multi-institutional, multi-PI approach, we are trying to understand this from different angles covering transcriptional reprograming, epigenetic reprograming, metabolic reprograming and immune modulators. My lab focuses on the transcriptional reprograming.
Research Programs
TIF1γ (TRIM33), an E3 ubiquitin ligase, is a negative regulator of TGF-β signaling with its well-established role in hematopoiesis. In cancer context, it is considered as a tumor suppressor with its negative regulatory role on TGF-β , which promotes tumor progression. In our lab we have shown that TIF1γ regulates self-renewal ability of oral cancer cells and causes the recurrence of the disease. We have unraveled its role in the transcriptional regulation of self-renewal genes. We have seen that TIF1γ is essential for the induction of self-renewal by many pathways. Currently, we are focusing on the upstream regulators of this pathway. We are also attempting to identify the post-translation modifications, like phosphorylation or acetylation that activates the molecule. Another important aspect to be explored is the role of the mutations of the gene that are prevalent in oral cancer patients.
CSCs reside in a special microenvironment called "CSC niche", maintained as a result of orchestration of different signaling pathways instigated through cell-cell interactions or secreted molecules. The niche helps in the maintenance of self-renewal ability, therapeutic resistance, and in turn regulate the relapse of the disease. We have been elucidating the role of EphA2/EphrinB1 signaling in CSC regulation in oral cancer. Our results implicate that the surface localization of the ligand and the receptor play a role in the regulation of oral cancer CSCs and recurrence. The mutations of these genes could be important in the modulation of this pathway.
The function of Host Defense Peptides (HDPs), an integral part of the innate immune system, has been recently reassigned to immunoregulation from the membranolytic antimicrobial activity. Contrary to the assumption that the antitumor activity of HDPs depends on their membranolytic activity, we observed that HDP-induced signaling through immunomodulators leads to apoptosis in cancer cells. We established that the peptide binds to IL6/IL6R/gp130 complex to modulate its downstream signaling. In contrast to the IL6 blockers that inhibit JAK/STAT activity, SSTP1 shifts the proliferative IL6/JAK/STAT signaling to the apoptotic IL6/JNK/AP1 pathway. Further, we show that cancer cells expressing high IL6Rα levels, like triple-negative breast cancer (TNBC), undergo apoptosis in response to low concentrations of SSTP1, which do not induce hemolysis, the result of the nonspecific membranolytic activity. Thus, our study highlights the importance of HDPs with identified targets for translation to clinical use.
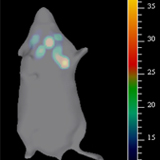
Single cell biology by and large depends on the detection and isolation of subpopulations from heterogeneous cell populations. Even though antibodies are conventionally used for this purpose, their relevance in certain context is limited, especially when the target is an intracellular molecule. For the first time we have shown a simple, cost-effective and efficient method of live sorting of cells based on the expression of an intracellular marker using a fluorophore-tagged binding peptide. Our results prove that our peptide TM2 can be used to separate cells with differential expression of HIRA.
Fluorescent-based optical imaging using Near Infrared (NIR) dyes tagged to tumor specific target is an optimal tool for surgical margin prediction. We have shown that TM1-IR680, is ideal for surgical margin prediction, in a preclinical analysis. Interestingly, the peptide was sensitive enough to detect lymph nodes that harbored dispersed tumor cells before colonization, which was impossible to identify by conventional histopathology.
The biology of oral cancer can be studied using in vitro and in vivo mouse models. We have successfully generated 3D co-culture model on alginate matrix where we could incorporate tumor fibroblasts and tumor cells. We are in the process of generating organoid model for oral cancer, which can be effectively used for drug screening. We have also established the generation of xenografts, orthotopic models and humanized orthotopic PDX models.
Current Research Grants
-
2026 2023
-
2025 2022
Differential Inhibition of Visfatin-PAK4 as a Novel Strategy in Esophageal Squamous Cell Carcinoma for Therapeutic Purpose (Co-PI).
Indian Council of Medical Research
-
2024 2021
Exploration of the role of fodrin, a protein required in functional microtubule reorganization, in cancer and apoptosis (Co-PI).
Indian Council of Medical Research
Previous/ Completed Research Grants
-
Virtual National Oral Cancer Institute Development of Animal Model Systems to study oral cancer progression
Department of Biotechnology [DBT] 2018-2022Evaluation of the role of TIF1γ in the regulation of self-renewal ability of OSCC stem cells
Department of Science & Technology [DST] 2016-2019Characterizing the signaling network that sustains oral cancer stem cells for developing a targeted therapy
Department of Science and Technology 2011-2014Identifying surface marker signature of oral cancer stem cells to develop a prognosis marker for oral squamous cell carcinoma using peptide-based CSC detection
Department of Biotechnology 2011-2014
Collaborations
-
Collaborations
1. Dr. Nebu Abraham George, Assistant Professor, Surgical Oncology, RCC, Thiruvananthapuram.
-
As a part of a multicentric project, VNOCI
1. Prof. Tapas K Kundu, Professor, Department of Molecular Biology and Genetics Unit, Jawaharlal Nehru Centre For Advanced Scientific Research, Bangalore
2. Dr. Annapoorni Rangarajan, Associate Professor, Molecular Reproduction, Development and Genetics, INDIAN INSTITUTE OF SCIENCE, Bangalore
3. Dr. Birendranath Banerjee, Associate Professor, School of Biotechnology, KIlT School of Bhubaneswar, Orissa
4. Dr. Debnath Pal, Associate Professor, Computational and Data Sciences, INDIAN INSTITUTE OF SCIENCE, Bangalore,Karnataka
5. Dr. Subhashini Sadasivam , National Centre For Biological Sciences, Bangalore
6. Prof. Anupam Chatterjee, Professor, Biotechnology & Bioinformatics, North-Eastern Hill University, Shillong
7. Prof. Partha Pratim Majumder, Distinguished Professor, Genomics, National Institute of Biomedical Genomics, West Bengal
8. Prof. Ramaswamy Subramanian, Senior Professor, Technologies for the Advancement of Science (TAS), National Centre For Biological Sciences, Bangalore
9. Prof. Saumyadipta pyne, Professor, Indian Institute of Public Health, Hyderabad
